Signaling dynamics in live single cells
A fundamental property of living cells is their extraordinary ability to sense and properly respond to a changing environment. Through highly interconnected networks of signaling proteins, internal and external cues are constantly evaluated to adjust cellular behavior. This ability is specially remarkable if one considers that signaling networks operate in the context of high macromolecular crowding, variable protein copy numbers, and are ultimately governed by the stochastic nature of molecular interactions. Through genetic and biochemical approaches coupled with powerful fluorescent biosensors, live cell imaging, and single molecule approaches, we explore and define new biological mechanisms that govern cell signaling.
We use as a model the Mitogen Activated Protein Kinase (MAPK) system, a widely conserved network of kinases that is involved in virtually all aspects of cell biology. Dysregulation of this multilayered network of kinases lies at the core of some of the most devastating human diseases including cancer, autoimmunity, and neurodegeneration. While MAPK cascades are among the most studied signaling pathways, two key gaps in our understanding of the network remain:
- The mechanisms of activation of the majority of MAP3Ks is unknown.
- How MAPK activities impact so many aspects of biology is unclear.
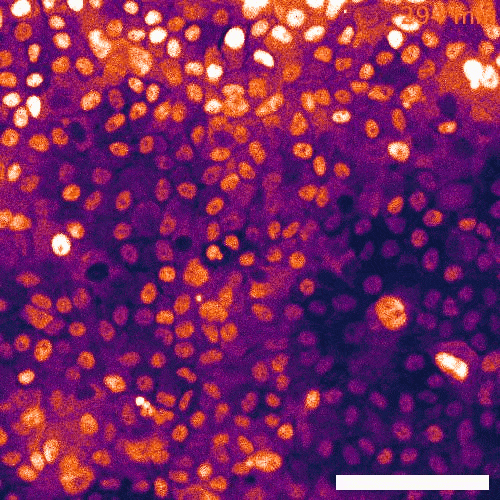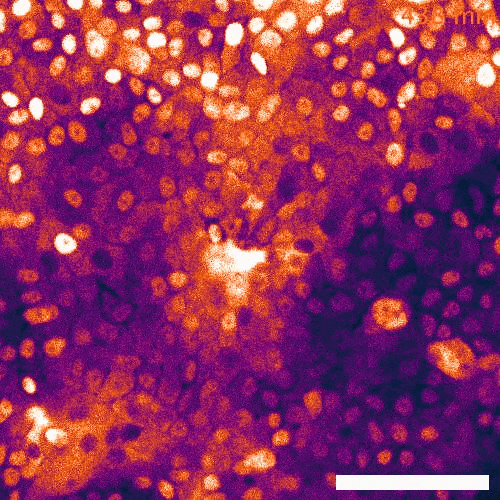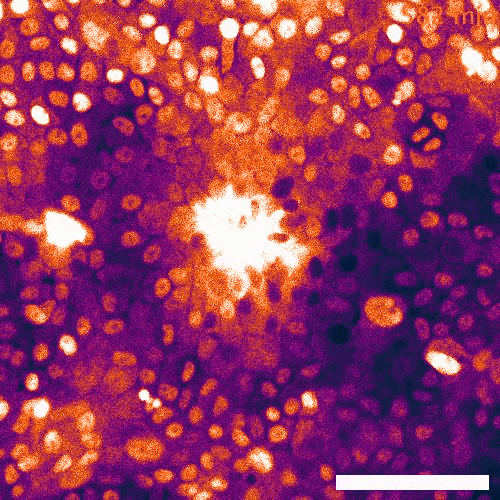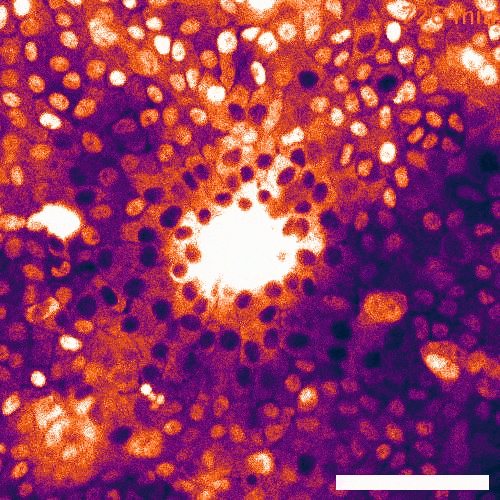
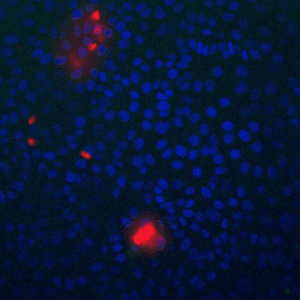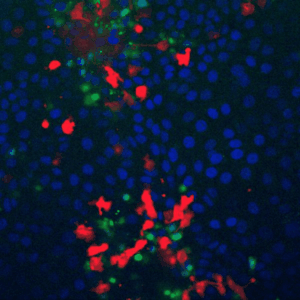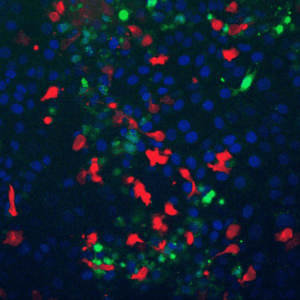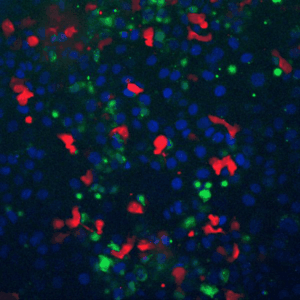
To fill these knowledge gaps, we combine single cell and single molecule live imaging approaches with gene editing and mouse models to enable rigorous examination of the physiological functions of the MAPK signaling network. We have developed new biosensors and fully automated imaging platforms to monitor signaling dynamics in single cells, unperturbed mouse tissues, and developing embryos. Using these approaches we have uncovered spatial and temporal patterns of signaling events that filter biological noise to effect deterministic outcomes. Our work has opened exciting new directions that suggest that the MAPK network is broadly responsible for monitoring and adjusting the biophysical properties of the cell including macromolecular crowding, phase transitions, and the mechanochemical properties of the cytoskeleton.