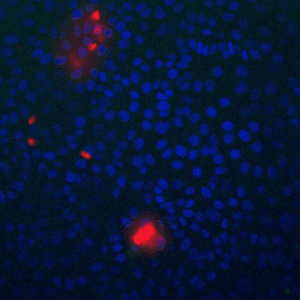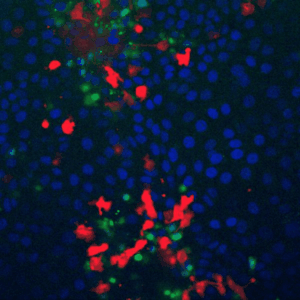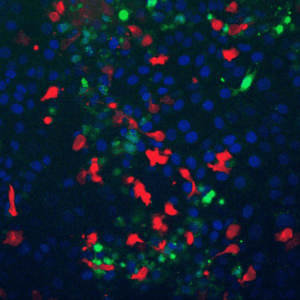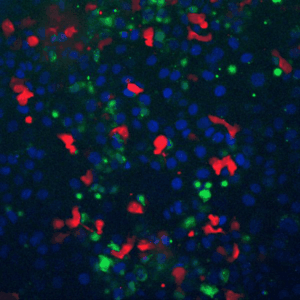
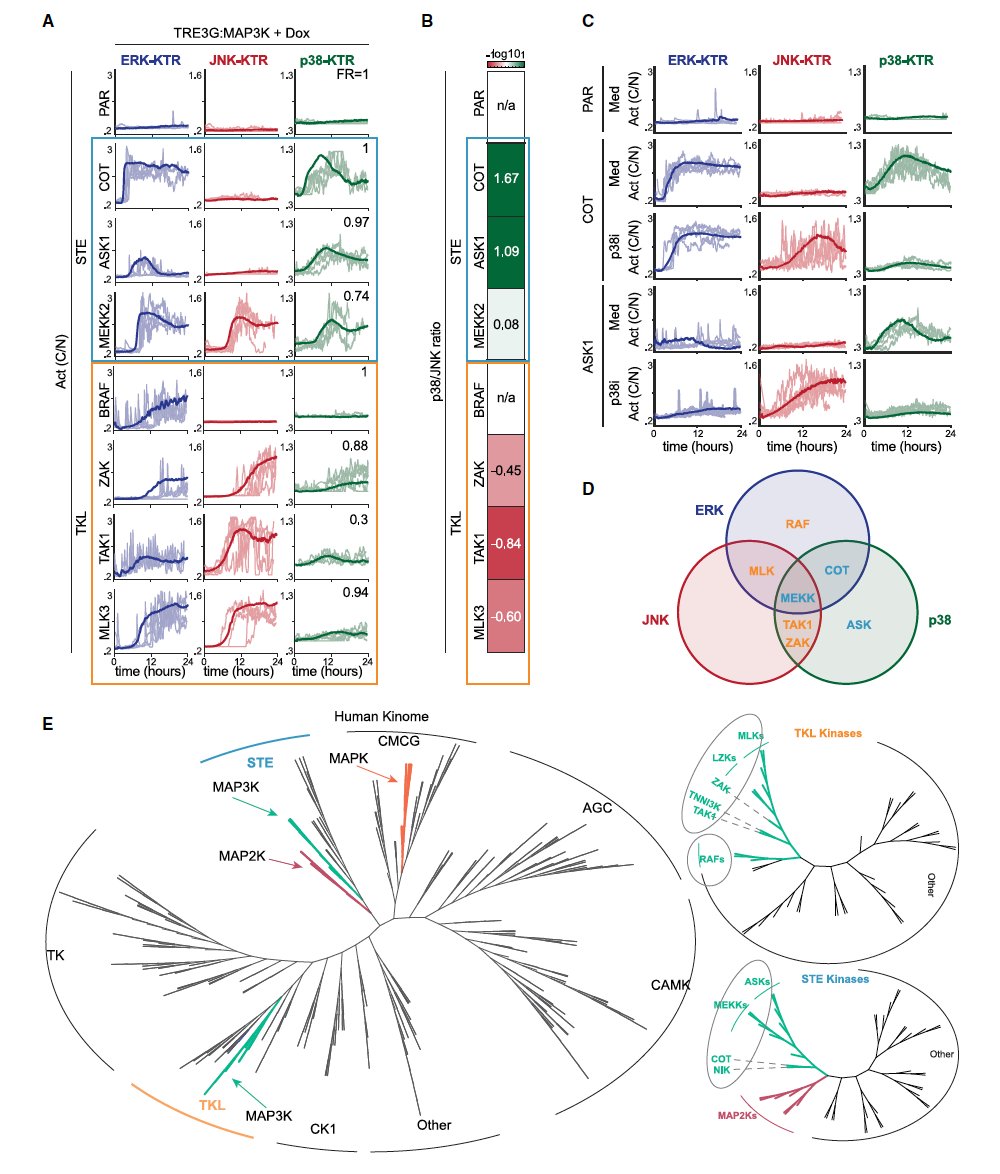
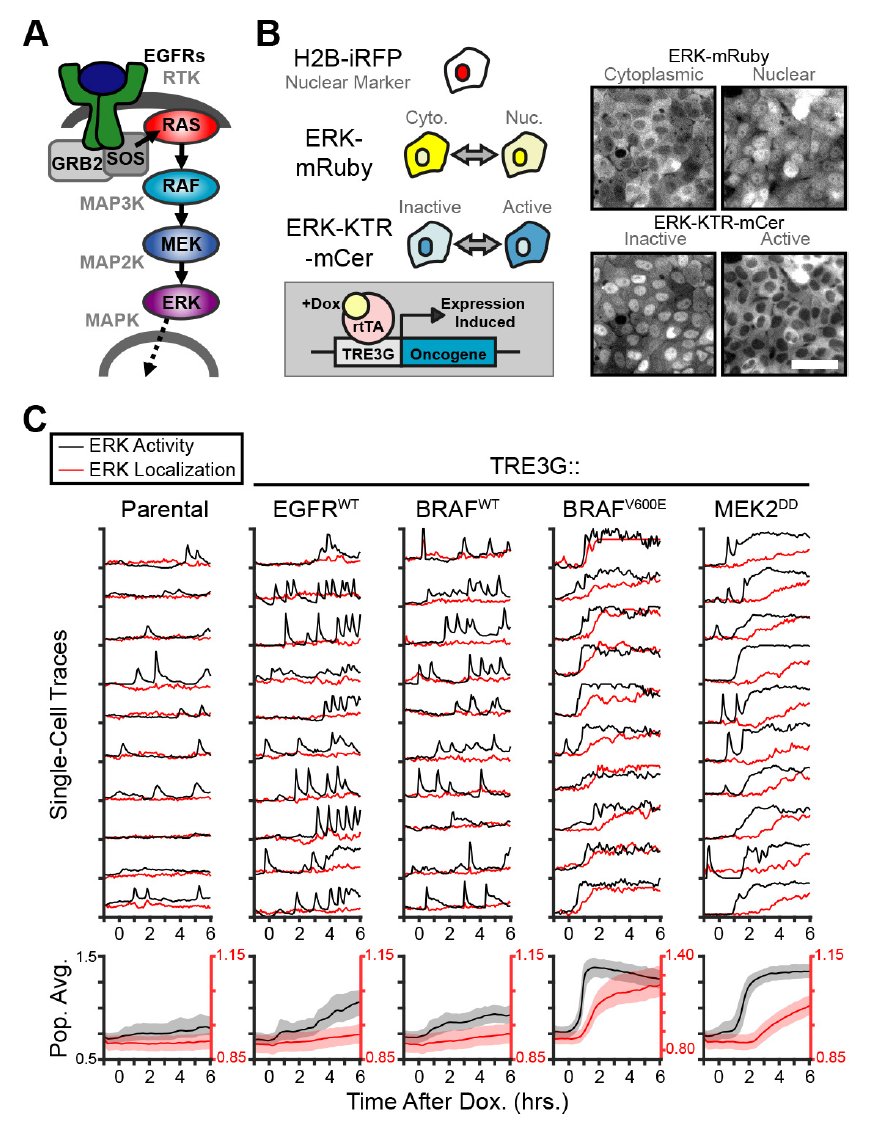
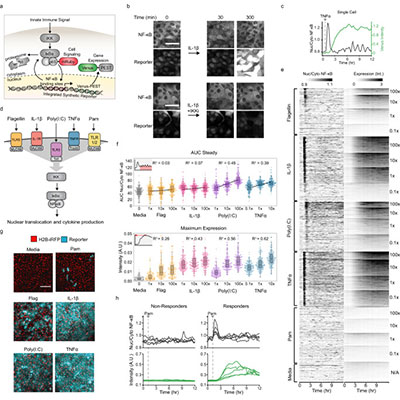
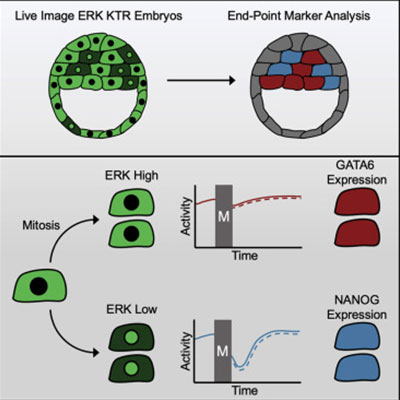
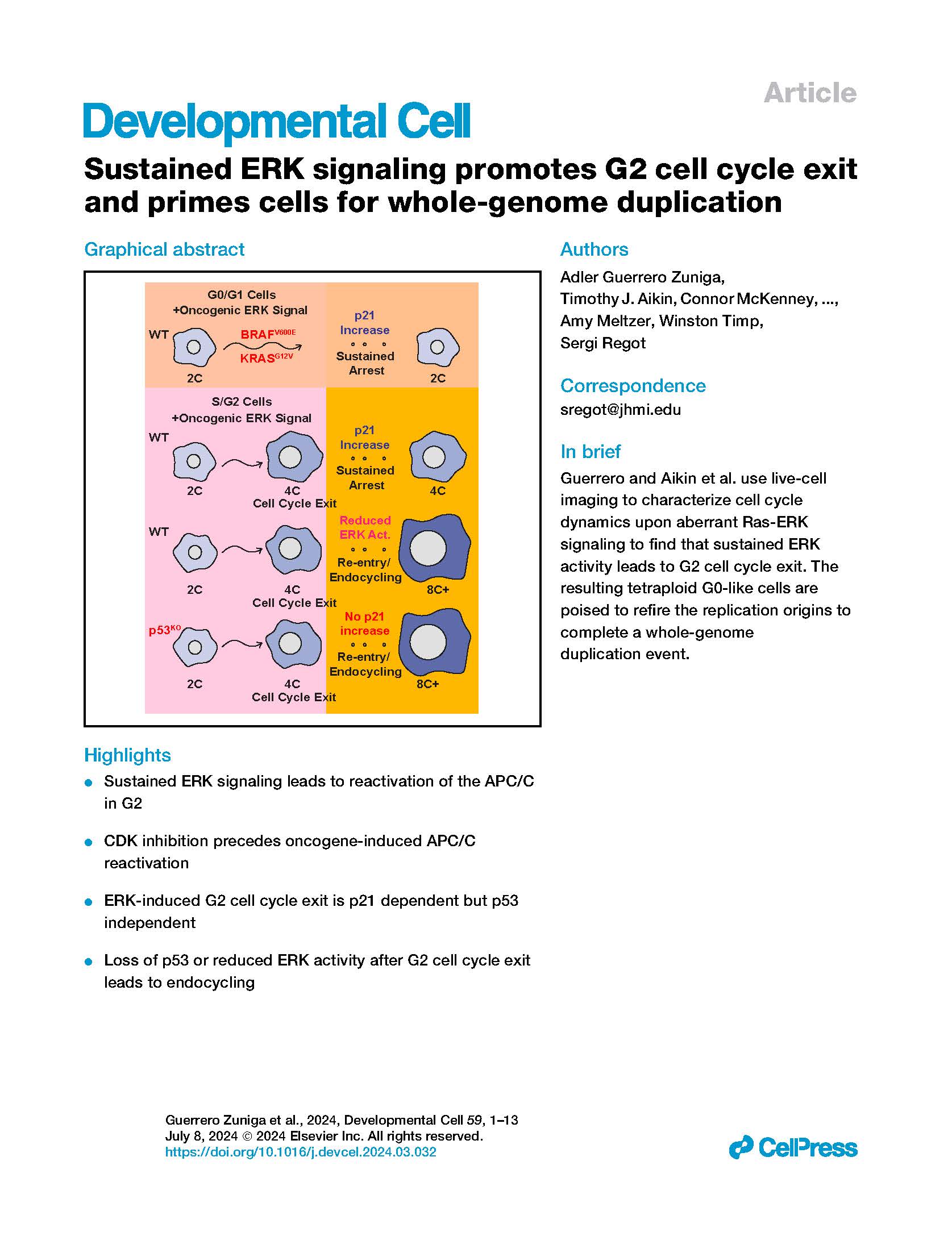
Cells live in a constantly changing environment, accordingly signaling networks have evolved to be extremely dynamic. To study signaling dynamics and its role in controlling cell cycle we use live cell imaging of fluorescent biosensors. These technologies provide the spatial and temporal resolutions required to understand how single cells sense and properly respond appropriately to a changing environment.
https://www.cell.com/developmental-cell/fulltext/S1534-5807(24)00200-4... Read More